@rdmpage@AlexHardisty@proibiosphere Well, take part in the process of clarification!
— Pensoft Publishers (@Pensoft) June 10, 2014I've been involved in a few Twitter exchanges about the upcoming pro-iBiosphere meeting regarding the "Open Biodiversity Knowledge Management System (OBKMS)", which is the topic of the meeting. Because for the life of me I can't find an explanation of what "Open Biodiversity Knowledge Management System" is, other than vague generalities and appeals to the magic pixie dust that is "Linked Open Data" and "RDF", I've been grumbling away on Twitter.
So, here's my take on what needs to be done. Fundamentally, if we are going to link biodiversity information together we need to build a network. What we have (for the most part) at the moment is a bunch of nodes (which you can think of as data providers such as natural history collections, databases, etc., or different kinds of data, such as names, publications, sequences, specimens, etc.).
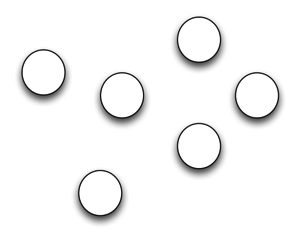
We'd like a network, so that we can link information together, perhaps to discover new knowledge, to serve as a pathway for analyses that combine different sorts of data, and so on:
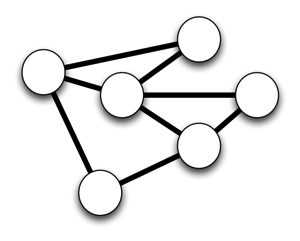
A network has nodes and links. Without the links there's no network. The fundamental problem as I see it is that we have nodes that have clear stakeholders (e.g., individual, museums, herbaria, publishers, database owners, etc.). They often build links, but they are typically incomplete (they don't link to everything that is relevant), and transitory (there's no mechanism to facilitate persistence of the links). There is no stakeholder for whom the links are most important. So, we have this:
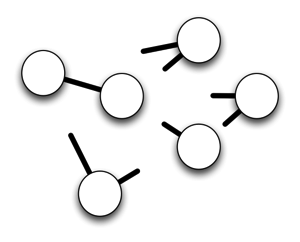
This sucks. I think we need an entity, a project, and organisation, whatever you want to call it for whom the network is everything. In other words, they see the world like this:
If this is how you view the world, then your aim is to build that network. You live or die based on the performance of that network. You make sure the links exist, they are discoverable, and that they persist. You don't have the same interests as the nodes, but clearly you need to provide value to them because they are the endpoints of your links. But you also have users who don't need the nodes per see, they need the network.
If you buy this, then you need to think about how to grow the network. Are there network effects that you can leverage, in the same way CrossRef has with publishers submitting lists of literature cited linked to DOIs, or in social media where you give access to your list of contacts to build your social graph?
If the network is the goal, you don't just think "let's just stick HTTP URLs on everything and it will all be good". You can think like that if you are a node, because if the links die you can still persist (you'll still have people visiting your own web site). But if you are a network and the links die, you are in big trouble. So you develop ways to make the network robust. This is one reason why CrossRef uses an identifier based on indirection, it makes it easier to ensure the network persists in the face of change in how the nodes serve their data. What is often missed is that this also frees up the nodes, because they don't need to commit to serving a given URL in perpetuity, indirections shields them from this.
In order to serve users of the network, you want to ensure you can satisfy their needs rapidly. This leads to things like caching links and basic data about the end points of those links (think how Google caches the contents of web pages so if the site is offline you may still find what you are looking for).
If your business depends on the network, then you need to think how you can create incentives for nodes to join. For example, what services can you offer them that make you invaluable to the nodes? Once you crack that, then all sorts of things can happen. Take structured markup as an example. Google is driving this on the web using schema.org. If you want to be properly indexed by Google, and have Google display your content in a rich form (e.g., thumbnails, review ratings, location, etc.) you need to mark up your page in a way Google understands. Given that some businesses live or die based on their Google ranking, there's a strong incentive for web sites to adopt this markup. There's a strong incentive for Google to encourage markup so that it can provide informative results for its users (otherwise they might rely on "social search" via Facebook and mobile apps). This is the kind of thing you want the network to aim for.
In summary, this is my take on where we are at in biodiversity informatics. The challenge is that the organisations in the room discussing this are typically all nodes, and I'd argue that by definition they aren't in a position to solve the problem. You need to pivot (ghastly word) and think about it from the perspective of the network. Imagine you were to form a company whose mission was to build that network. How would you do it, how would you convince the nodes to engage, what value would you offer them, what value would you offer users of the network? If we start thinking along those lines, then I think we can make progress.
 It is almost a year to the day that I
It is almost a year to the day that I