Baldwin, C. C., & Smith, W. L. (1998). Belonoperca pylei, a new species of seabass (Teleostei: Serranidae: Epinephelinae: Diploprionini) from the cook islands with comments on relationships among diploprionins. Ichthyological Research, 45(4), 325–339. doi:10.1007/BF02725185
After extracting some data from ZooBank API I created a DOT file connecting the various "taxon name usages" associated with Belonoperca pylei and constructed a graph using GraphViz:
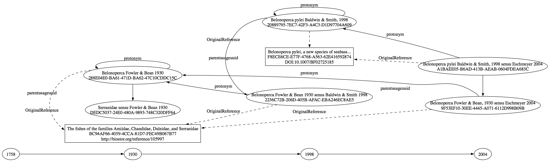
You can grab the DOT file here, and a bigger version of the image is on Flickr. I've labelled taxon names and references with plain text as well as the UUIDs that serve as identifiers in ZooBank. (Update: the original diagram had Belonoperca pylei Baldwin & Smith, 1998 sensu Eschmeyer [9F53EF10-30EE-4445-A071-6112D998B09B] in the wrong place, which I've now fixed.)
This is a fairly simple case of a single species, but it's already starting to look a tad complicated. We have Belonoperca pylei Baldwin & Smith, 1998 linked to its original description (doi:10.1007/BF02725185) and to the genus Belonoperca Fowler & Bean, 1930 (linked to its original publication http://biostor.org/reference/105997) as interpreted by ("sensu") Baldwin & Smith, 1998. Belonoperca Fowler & Bean 1930 sensu Baldwin & Smith 1998 is linked to the original use of that genus (i.e., Belonoperca Fowler & Bean, 1930). Then we have the species Belonoperca pylei Baldwin & Smith, 1998 as understood in Eschmeyer's 2004 checklist.
Notice that each usage of a taxon name gets linked back to a previous usage, and names are linked to higher names in a taxonomic hierarchy. When the species Belonoperca pylei was described it was placed in the genus Belonoperca, when Belonoperca was described it was placed in the family Serranidae, and so on.
 Quick note on an experimental version of
Quick note on an experimental version of 
 Benoît Fontaine et al. recently published a study concluding that average lag time between a species being discovered and subsequently described is 21 years.
Benoît Fontaine et al. recently published a study concluding that average lag time between a species being discovered and subsequently described is 21 years.